Blog
Scientists follow Leprosya roots in South America at the back of 4000 years
In the last study published in the journal Scientists analyzed the genomes of 4,000 years of human remains, revealing a long leprosy history in the Americas.
Linden (also known as Hansen’s disease) is caused and. He was recognized as the second causal pathogen of Hansen’s disease and is associated with more severe forms of the disease, including the phenomenon of Lucio and dispersed leprosy leprosy. Unrepected people can develop chronic peripheral neuropathy and physical impairment. Many infected people remain asymptomatic, hindering diagnostic and control measures.
Analyzes of ancient genome confirm his contagious potential lasting millennium. People are the main host of Hansen’s disease, but maintaining causal bacteria in animals raises concerns about their potential as animal tanks. Nine armored bands are known sources, and red squirrels can contain both and. A recent detection in the archaeological bone of rodents suggests infectiousness between species in historical periods.
Understanding the evolutionary history and distribution is limited because only a few cases of infection have been molecularly confirmed. Research suggests his presence in Southeast Asia and America. Analyzes covering ancient and modern genomic data consistently support origin outside America. However, detection was not reported in archaeological contexts.
Examination and results
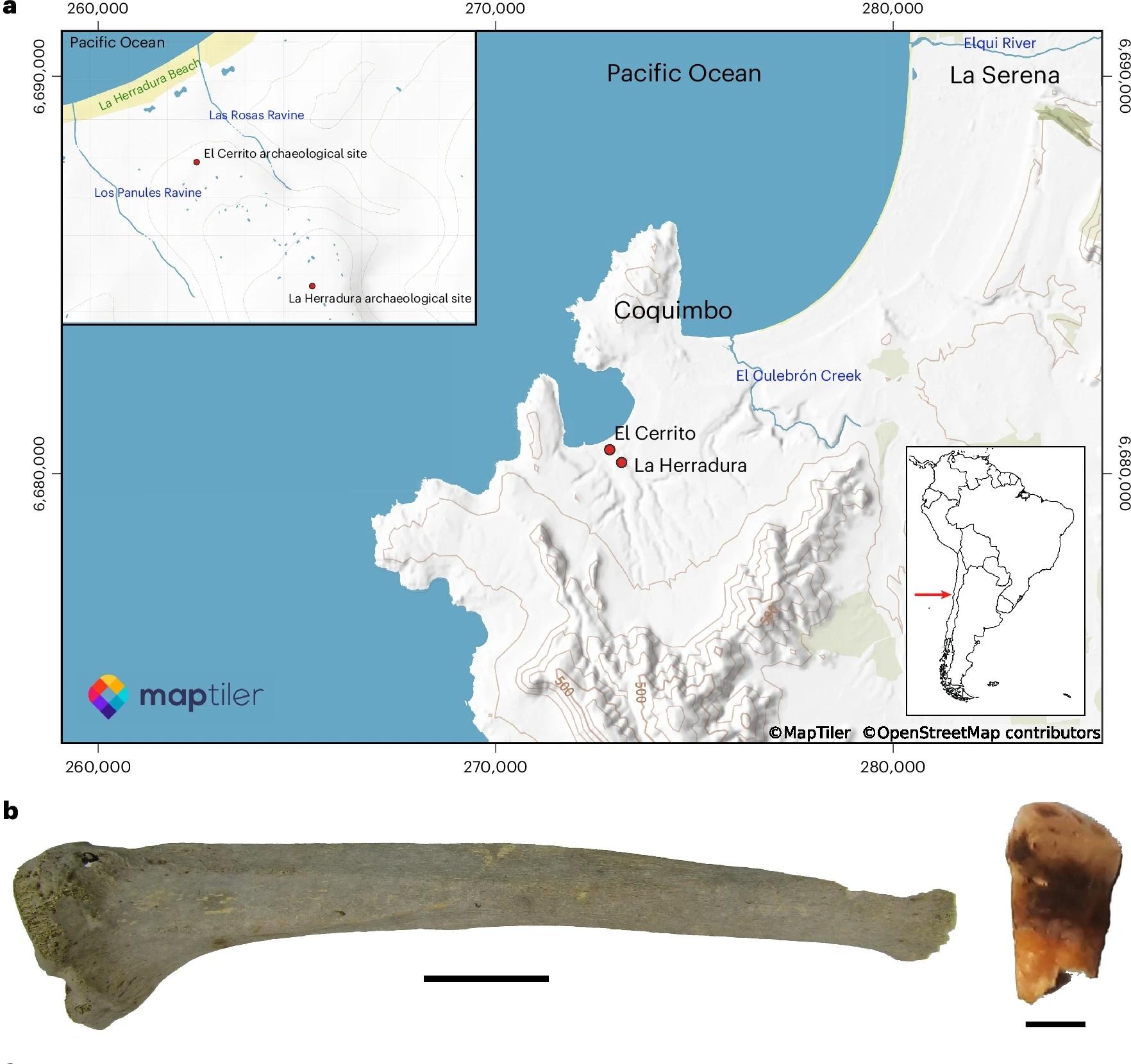
In this study, scientists analyzed the genomes of 4,000-year-old human skeletal remains from various archaeological contexts. First 19 bones and 35 teeth with pathological changes suggesting an infection, belonging to 41 people, samples from five archaeological places in Chile were taken.
The article notes that although bone changes in two infected people were in line with Hansen’s disease, they themselves were not definitively diagnostic, emphasizing the importance of molecular analysis for confirmation. A small amount of each tissue was extracted and constructed by the DNA library for sequencing.
The data was examined in terms of various pathogenic viruses and bacteria in accordance with the hypothesis free method. This revealed several thousand DNA fragments with homology in two archaeological tissues, a man of a man, called ECR003 in the place of El Cerrito and Tibbles from another man (ECR001) in the place of La Herradura. Radiocarbon dating of these two elements indicated that they were contemporary from 3900 to 4100 years ago.
DNA libraries were enriched using a probe set designed from a modern reference panel, which caused some uneven cover, but still gave extremely high quality ancient genomes. These libraries were then sequentized to examine the suitability of genomic reconstruction. Various species of mycobacteria were distinguished using a competitive approach to mapping. The average genomic coverage was 74 times for ECR003 and 45 times for ECR001, when it was mapped to the modern reference of the genome isolated from the Mexican patient.
In addition, scientists have examined the discrepancy between the genomic distribution and the reduction in relation to evolutionary time scales. To this end, the pumping analysis revealed a high level of discrepancy, with about half of the protein encoding regions showing at least 50% of the sequence homology between two pathogens. The mapping approach showed that two pathogens had only ~ 25% of nucleotide identity.
Then the relationship was examined between and other mycobacterial pathogens, analyzing Locus RNA RNA 16S. This indicated that this is the closest relative, despite their discrepancies.
In addition, a conservative phylogenetic reconstruction was carried out at the genome level, focusing on diversity inside and was limited to two ancient genoms, four contemporary human genome and six modern red squirrel genome.
There was a solid and clear separation of lines related to rodents and a man in which the ancient genomes formed a sister claw to a group of all human sequences.
Comparative analysis of the study also undermined previously reported genes from India, which suggests by competitive mapping, which showed a much larger homology.
In addition, time -generated phylogenetic trees have been generated using the radiocarbon centuries of the ECR001 and ECR003 skeletal elements, along with the year of collecting all modern genomes to estimate the indicators of evolution and times of discrepancy.
The evolutionary indicator was estimated at 6.91 x 10-9 Replacing a place annually, adapting to estimates. The median time for the latest common ancestor (TMRCA) was estimated about 26,800 years ago; However, the authors note that a small number of available genomes causes a wide potential range of date from 4206 to 115 340 years ago.
The divergence time for genomes from human hosts was estimated at about 12,600 years (with a range from 5304 to 49,659 years ago), while TMRCA for the red squirrel klade was much newer 440 years.
Conclusions
To sum up, the study reveals a clear evolutionary history. This deep timeline, potentially reaching for the transition of Pleistocene-Holocene, contrasts sharply with other main pathogens, such as (plague bacteria), which, as it is believed, appeared recently in the neolithic era along with the growth of agriculture.
The fact that infections took place in South America before the periods of known contact with European populations or oceans imply the movement of pathogens in human groups during early population or endemicity on the continent in another species of the reservoir. The latter suggests that his current distribution comes from postcolonial distribution, which makes him one of the few diseases that we know that they appeared in the Americas.
The article defines these discoveries in the perspective of “one health”, calling for a broader supervision of animal tanks in order to better understand the ecology of the disease and the potential of the animal.
Reference to the journal:
- Ramirez da, sitter tl, Översti s, and others. 4000-year-old Genomy from Chile reveal a long establishment of Hansen’s disease in the Americas. , 2025. DOI: 10.1038/S41559-025-02771-Y, https://www.nature.com/articles/s41559-025-02771-y